AI-Powered Retinal Disease Detection with Deep Learning & Computer Vision Pipeline
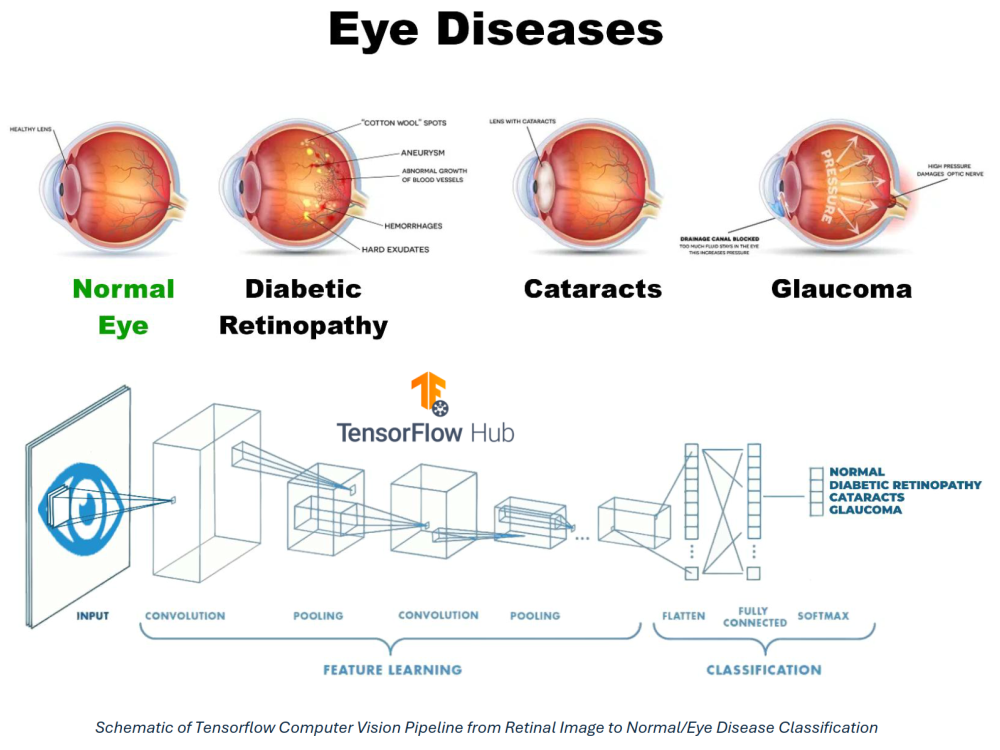
This project developed a deep learning–based computer vision pipeline to automatically classify retinal fundus images into four categories: cataract, diabetic retinopathy, glaucoma, and normal. The goal is to demonstrate how AI can support early screening by flagging at-risk patients for follow-up with ophthalmologists.
Using transfer learning with an EfficientNet-based CNN, the pipeline ingests high-resolution retinal images, performs preprocessing and data augmentation, and trains a multi-class classifier with performance tracking across training, validation, and test splits.
Background & Problem Statement
Diabetic eye diseases—including cataracts, diabetic retinopathy, and glaucoma—are leading causes of preventable blindness worldwide. Early detection through retinal fundus imaging can significantly improve clinical outcomes, but manual diagnosis is time-consuming, subjective, and requires specialized expertise that may not be available in low-resource settings.
Advances in deep learning and transfer learning have enabled convolutional neural networks (CNNs) to achieve near-expert performance on a variety of medical imaging tasks. However, deploying such models in a realistic workflow requires more than a single notebook: the full pipeline must handle data ingestion, preprocessing, training, evaluation, and model interpretability.
Problem Statement: Can we design an end-to-end computer vision pipeline that uses deep learning to classify retinal images into common disease categories and normal, in a way that is accurate, reproducible, and amenable to future clinical integration?
Model Development
The project focused on building a robust training pipeline with strong generalization and clear evaluation metrics.
- Dataset Preparation: Curated a dataset of over 4,000 retinal fundus images, each resized to 456×456 pixels to preserve fine-grained structural features (e.g., vessels, optic disc, lesions) while keeping training efficient.
- Train/Validation/Test Splits: Stratified the data into training, validation, and test sets to maintain class balance and enable unbiased performance estimates.
- Preprocessing & Augmentation: Applied standard computer vision preprocessing (resizing, normalization) along with augmentation such as random rotations, flips, zooms, and brightness/contrast adjustments to increase effective sample size and improve robustness to acquisition variability.
- Model Architecture (Transfer Learning): Used an EfficientNet-based backbone with ImageNet pre-trained weights, removing the final classification layer and adding a new multi-class head (dense layers with dropout and a softmax output) tailored to the four retinal classes.
- Training Strategy: Started by freezing the backbone and training only the new head, then progressively unfroze later EfficientNet blocks for fine-tuning with a lower learning rate. Monitored loss and accuracy on training and validation sets to avoid overfitting.
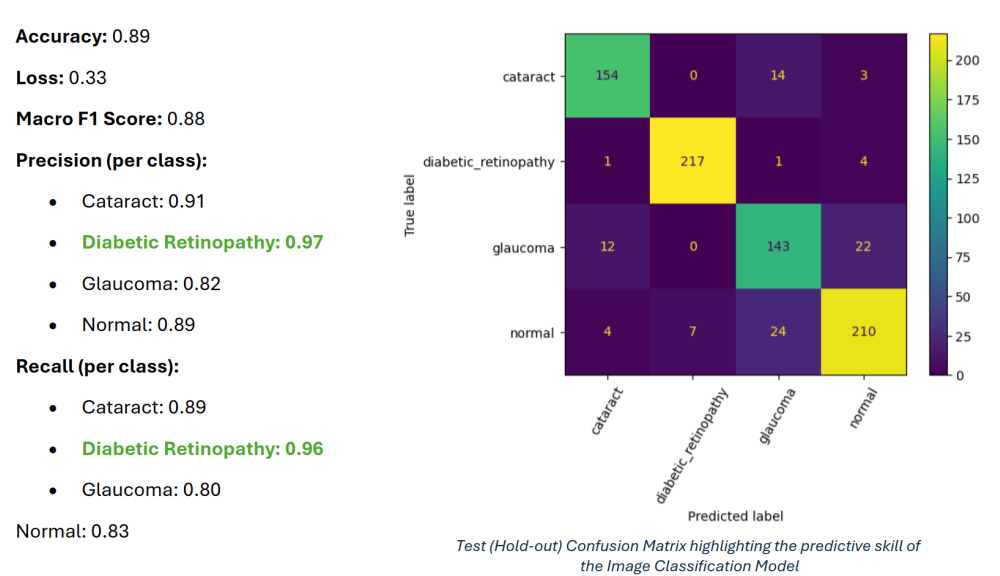
- Loss & Metrics: Optimized using categorical cross-entropy with accuracy and per-class precision/recall as key metrics. Tracked training via learning curves and confusion matrices.
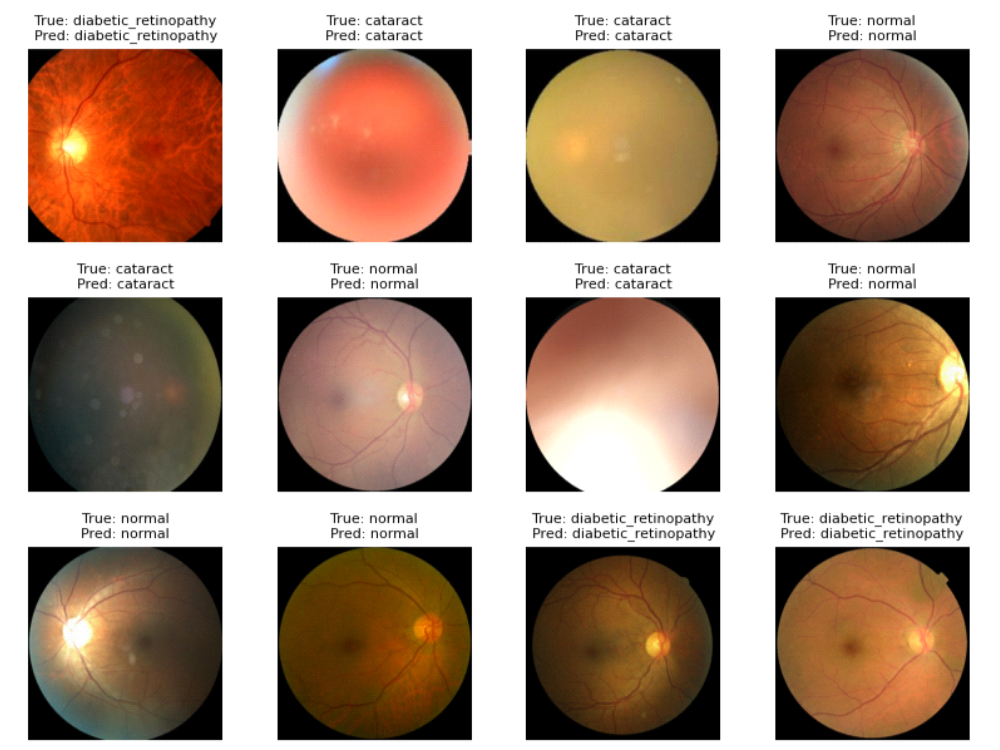
Computer Vision Pipeline & Evaluation
Beyond the core model, the project emphasizes a pipeline-oriented approach and basic visual explainability to support future clinical interpretation.
- End-to-End Pipeline: Wrapped data loading, preprocessing, model definition, training, and evaluation into a parameterized pipeline, making it straightforward to rerun experiments with different architectures, hyperparameters, or image resolutions.
- Evaluation on Held-Out Test Set: Assessed performance on a strictly held-out test set, computing overall accuracy as well as per-class precision, recall, and F1 scores to ensure that minority disease classes were not neglected.
- Class Imbalance Handling: Used class weights and augmentation to reduce bias toward majority classes and encourage the model to properly learn underrepresented categories (e.g., glaucoma vs. normal).
- Reproducibility: Encapsulated the experiment in a Jupyter notebook / script with explicit random seeds, dataset splits, and configuration blocks to make the workflow reproducible and extensible.
Impact & Actionable Insights
While this project is research-focused rather than deployed in a live clinic, it demonstrates a full-stack approach to medical imaging ML that can be adapted for real-world screening workflows.
- Screening Support: Shows how deep learning can assist clinicians by pre-screening retinal images, flagging potential cases of cataract, diabetic retinopathy, or glaucoma for further review.
- Pipeline Reusability: The codebase and pipeline design can be easily adapted to other medical imaging tasks (e.g., chest X-rays, dermoscopy) by swapping the dataset and potentially adjusting the backbone architecture.
- Model Governance Readiness: Structuring the work into well-documented steps (data, model, evaluation, explainability) aligns with emerging best practices for healthcare AI validation and governance.
- Portfolio Demonstration: Highlights experience with computer vision, transfer learning, class imbalance, and basic model interpretability—skills that translate directly to many real-world data science and ML roles, inside and outside healthcare.